Integrative Transcription Start Site Analysis and Physiological Phenotyping Reveal Torpor-specific Expressions in Mouse Skeletal Muscle
Genshiro A Sunagawa, Ruslan Deviatiiarov, Kiyomi Ishikawa, Guzel Gazizova, Oleg Gusev, Masayo Takahashi
doi: https://doi.org/10.1101/374975
We identified 287 torpor-specific promoters in muscles from torpid mice. Furthermore, we found that transcription factor Atf3 is up-regulated by torpor deprivation. These results are uploaded to bioRxiv. The preprint is submitted to a scientific journal to be reviewed for future publication.
Introduction
Mice enter an active hypometabolic state, called daily torpor, when they experience a lowered caloric intake under cool ambient temperature. We are interested in applying such active hypometabolism to medicine by understanding the mechanism of it. During torpor, the animal’s oxygen consumption rate drops to less than 30% of the normal rate, which cannot be achieved without hypometabolism in peripheral tissues. To understand the mechanism of metabolic suppression during torpor in peripheral tissues, we systematically analyzed the expression landscape of transcription start sites (TSS) in mouse skeletal muscles under various metabolic states and identified torpor-specific transcription patterns.
Two genetically close inbred strains, C57BL/6J (B6J) and C57BL/6N (B6N), show distinct torpor phenotypes.
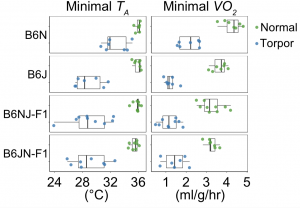 In 2016, we developed a reproducible method for inducing torpor in B6J (Sunagawa & Takahashi, 2016). Using this method, we found that B6N had a shallower torpor phenotype than B6J. Furthermore, the offspring of these two strains inherited the B6J-phenotype dominantly, clearly indicating the effect of genetic background on the torpor phenotype.
In 2016, we developed a reproducible method for inducing torpor in B6J (Sunagawa & Takahashi, 2016). Using this method, we found that B6N had a shallower torpor phenotype than B6J. Furthermore, the offspring of these two strains inherited the B6J-phenotype dominantly, clearly indicating the effect of genetic background on the torpor phenotype.
Systematic transcription start site (TSS) analysis identified 287 torpor-specific promoters in mouse skeletal muscle.
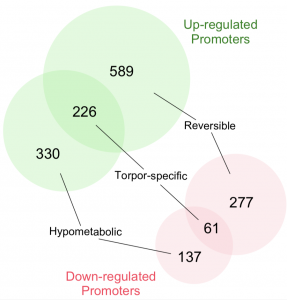 To understand what gene is differentially expressed in muscles during torpor, we used CAGE-seq to obtain the comprehensive TSS profile of a sample. Two sets of experiments were performed, testing the reversibility and the association with hypometabolism of the TSS profile in muscles with various metabolic conditions. We found 287 torpor-specific promoters (226 up- and 61 down-regulated).
To understand what gene is differentially expressed in muscles during torpor, we used CAGE-seq to obtain the comprehensive TSS profile of a sample. Two sets of experiments were performed, testing the reversibility and the association with hypometabolism of the TSS profile in muscles with various metabolic conditions. We found 287 torpor-specific promoters (226 up- and 61 down-regulated).
Transcription factor Atf3 is highly expressed during torpor deprivation.
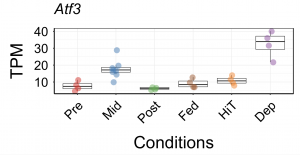 Torpor entrance can be easily bothered by gently touching the mouse body. Therefore, if we keep bothering the animal, it is possible to produce a torpor-deprived condition, which is a state that the animal cannot have enough torpor. We evaluated the TSS profile from muscles taken from torpor-deprived animals and identified Atf3 as an over-expressed gene, which was also one of the enriched motifs in the torpor-specific promoters.
Torpor entrance can be easily bothered by gently touching the mouse body. Therefore, if we keep bothering the animal, it is possible to produce a torpor-deprived condition, which is a state that the animal cannot have enough torpor. We evaluated the TSS profile from muscles taken from torpor-deprived animals and identified Atf3 as an over-expressed gene, which was also one of the enriched motifs in the torpor-specific promoters.
Conclusion
This study showed the usefulness of mice for active hypometabolism. It was possible because of the rich genetic information and state-of-art genetic technology in mice. Mice are also known to have a very powerful in vitro experiment environment, which is the cell/tissue culture systems. Therefore, we are planning to extend our hypometabolism research to in vitro assays using cultured mouse cells.
Awknolegments
This work was supported by the RIKEN Special Postdoctoral Researcher program and by Grant-in-Aid for Scientific Research on Innovative Areas (Thermal Biology) 18H04706 from MEXT.
Leave a Reply